Think about aphids — those little, green, garden-destroying bugs. The bane of the begonia. But aphids exist in an ecosystem. Which is to say, those little suckers have enemies. When gardeners see aphids, it’s often ladybugs to the rescue.
There is another plague out there right now: the antibiotic-resistant bacteria currently making a home in our health care system that kills thousands each year.
But by thinking like a gardener and looking to the ecosystem those microbes originally came from, we may be able to find new ways to fight them.
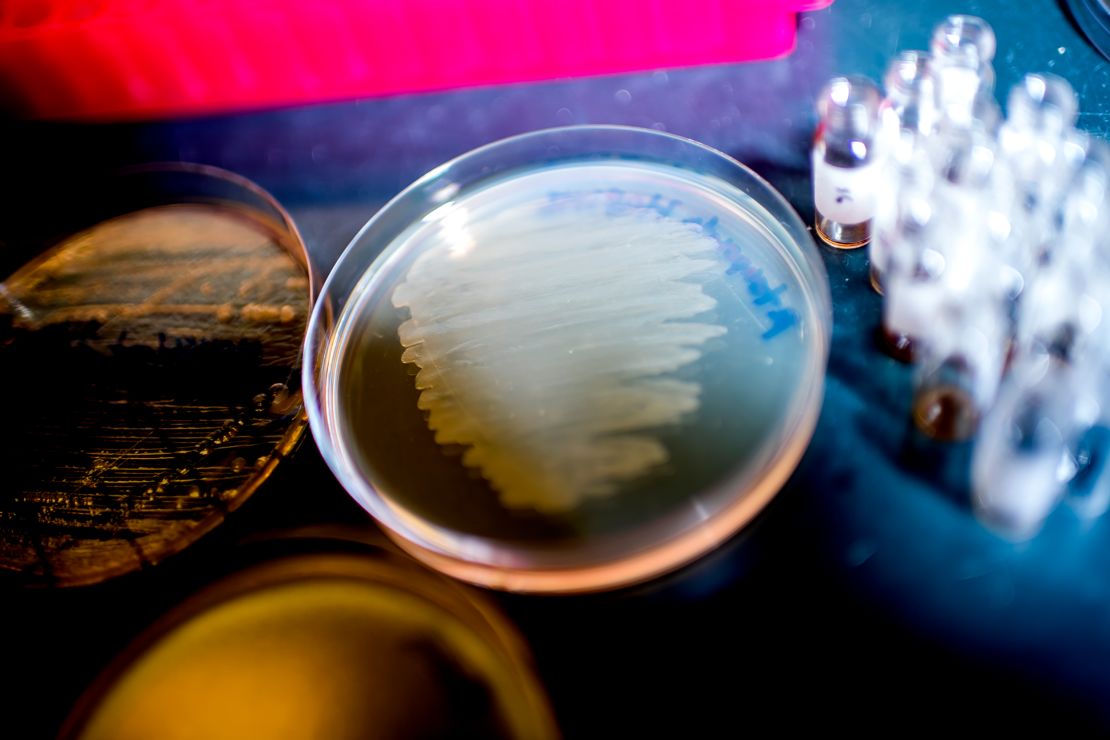
For instance, researchers at Vanderbilt University recently unearthed a new kind of chemical compound that may one day help fight off antibiotic-resistant bacteria. The source? Dirt.
Dirt, it turns out, contains an entire microbial ecosystem, with a wealth of intriguing compounds. And while there are major hurdles between discovering a new drug and using it, scientists are hopeful new techniques could pay off.
Stuck in the mud
Antibiotic-resistant microbes, commonly called superbugs, have become a major health problem across the world. More than 2.8 million infections happen every year in the United States alone; more than 35,000 people will die from those infections.
The answer for an infection is often antibiotics. Or stronger antibiotics.
But we’re not the original inventor of this class of compounds — for time immemorial, antibiotics have been deployed by the microbes themselves. Microbes, such as the myriad species of fungi, bacteria and other critters that live in the soil, live in a complex ecosystem where chemical defenses are often necessary in order to fend off or attack competitors.
In fact, soil was actually one of the great historical sources for new antibiotics.
“The vast majority of antibiotics we use today come from growing bacteria out of soil,” said Sean Brady, a chemical biologist and professor at The Rockefeller University in New York City. Though we came up with ways to make them ourselves, it was in dirt that these antibiotics were first discovered.
Streptomycin, which is often used to treat tuberculosis, came from a sample of New Jersey soil back in the 1940s, for instance.
At the time, a wealth of new medicines seemed just within reach.
Unfortunately, it turned out that soil microbes are finicky. Only a handful grow well in traditional laboratory setups, and what we could get out of them was limited. Stuck discovering the same compounds over and over again, the pharmaceutical industry’s focus shifted to tweaking the antibiotics we already had.
Returning to our roots
However, in the last few years, a number of researchers have been going back to the soil with new techniques.
One potential breakthrough came when Kim Lewis, a microbiologist and university distinguished professor at Northeastern University, and his colleague Slava Epstein, a professor of biology, figured out how to “domesticate” wild microbes. The duo loaded soil bacteria into a special device called the “iChip” (short for isolation chip), then buried it back into their native soil until the bacteria grew into usable colonies.
One compound Lewis and Epstein’s team discovered, called teixobactin, originally came from a grassy field in Maine. Teixobactin got a write-up in Nature in 2015 and was hailed as the first truly novel antibiotic to be discovered in nearly 30 years.
Brady and his team announced a few years later that they had managed to discover a class of compounds they called malacidins, again from soil samples. In this case, the researchers skipped growing finicky bacteria altogether, instead processing bits of the bacterial genome from the soil directly, a technique known as metagenomics.
These discoveries, and others like them, could represent major breakthroughs in fighting antibiotic-resistant bacteria.
Neither teixobactin nor malacidins are ready for use yet, though.
Teixobactin is currently being studied by a company called NovoBiotic Pharmaceuticals, which said in an email statement that the compound is in preclinical development, with the goal to submit it for FDA approval to begin testing in humans in 2022.
Brady, meanwhile, said his research team is continuing to investigate the malacidin family of compounds and are hoping to narrow down the candidates to the ones that would have the best chance of working in humans before moving toward trials.
“You don’t always want to go with the very first thing you find,” he said.
The truth is, for all their potential, these drugs may still fail. Both Lewis and Brady, as well as industry experts, caution that the pipeline for turning these discoveries into drugs can be long and, it turns out, pretty leaky.
The leaky pipeline
“It’s exciting to have these potential new novel classes of drugs, but there’s a lot of work that needs to be done to take it from soil to patient,” said Wes Kim, a drug development expert and senior officer at Pew Charitable Trust’s Antibiotic Resistance Program.
The trust has been advocating and supporting the development of new antibiotics for a long time. The nonprofit institution is committed to seeing these kinds of discoveries come to light.
But developing any new drug, not just antibiotics, can be time-consuming and difficult. Drugs that look promising in petri dishes may turn out to be ineffective in the real world. Others may have side effects so severe they’re not worth using. Overall, only about one in every five infectious disease drugs that reach human testing end up getting FDA approval.
And all this testing takes time and money. Estimates for the average cost of drug development start in the hundreds of millions of dollars.
Antibiotics, it turns out, may not make enough to pay off this risk. Counterintuitively, despite the growing need for new antibiotics in general, individual antibiotics themselves don’t actually make a lot of money.
This can be for a host of reasons. Antibiotic prescriptions tend to be short, for instance. Hospitals may also try to hold back on using new antibiotics as well, saving them for dire situations.
This comparatively low return on investment means many large pharmaceutical companies have left the field. Nearly all the products in development today are being studied by small companies, which may struggle with the financial and logistical hurdles of long clinical trials and marketing.
There are potential fixes, though. The Generating Antibiotic Incentives Now, or GAIN, Act of 2012, for example, sought to create financial incentives for companies, including more time holding the exclusive rights to a drug. The as-of-yet-unpassed Developing an Innovative Strategy for Antimicrobial Resistant Microorganisms Act, or DISARM Act, would tweak Medicare rules to incentivize hospitals to use new drugs.
Another key fix would be compensating for the leaky pipeline by simply increasing the number of new candidate drugs being discovered.
“The bottleneck by consensus is clearly in discovery,” Lewis said. “You need lots of compounds to start with because of attrition during development.”
And that means taking those amazing, newsworthy breakthroughs and doing them again. Not just once, but over and over again.
Digging far and wide
Tucked away in a closet of Brady’s lab are boxes and boxes of dirt from all around the country. Each shoebox-size container contains a number of samples — some from fellow academics, but many the end result of a crowdsourcing campaign that asked citizen science volunteers to go prospecting themselves.
His lab isn’t the only group that has taken advantage of crowdsourcing. The Small World Initiative, which uses the study of soil microbes and antibiotic resistance to help educate students, has also collected a wealth of samples the program hopes will contribute to the pipeline.
“Students are engaged by looking into something that solves a real challenge,” said Erika Kurt, the group’s CEO. “And since there are so many new things to find in soil, every single student finds something that’s exciting.”
Researchers are also looking to take these all-important new methods and use them more and more often. Brady and his team, for instance, are continuing to expand their use of metagenomics; researchers elsewhere are also using this approach to look in other places, like ocean water.
“The technologies are beginning to catch up with the ideas,” Brady said. Hopefully over the next decade, these methods will deliver steadily more potential candidates. “We’ve already started to see it and we’re going to see even more of it over the next decade,” he said.
The iChip — that device that Lewis’ team used to “domesticate” wild bacteria — has seen steady use as well, though Lewis is also looking further afield. His team also recently discovered a compound called darobactin in the guts of soil nematodes, for instance.
It’s potentially very exciting, he said, because it can affect what’s known as gram-negative bacteria, which includes notable bugs like E. coli and Salmonella. Lewis called this discovery the “Holy Grail” due to how notoriously difficult gram-negative bacteria can be to treat.
Get CNN Health's weekly newsletter
Sign up here to get The Results Are In with Dr. Sanjay Gupta every Tuesday from the CNN Health team.
It’s possible that none of these new compounds will end up making it all the way into doctors’ hands, of course, but the more novel compounds we can find, the more likely it is that one will.
“The more we can increase our shots on goal, so to speak, I think the better we are,” Kim said.
James Gaines is a Seattle-based freelance science journalist who specializes in environment and solutions stories.